Drug Repurposing
Despite technological advances, the development of therapeutic discoveries proceeded much slower than expected. Due to the relatively low risk of failure in early-stage trials, development costs and time advantages, drug repurposing have attracted attention in drug discovery.
In recent study, the costs of repurposing drug to market have been estimated to be US$300 millon on average, compared with an estimated ~$2-3 billion for a NCE (New Chemcal Entity). (Nature, 2016)
Historically, drug repurposing has been opportunistic and coincidence rather than systematic investigation, as illustrated by sildenafil citrate (Viagra, pfizer). It held a market-leading 47 % share of the erectile dysfunction in 2012, with worldwide sales totalling $2.05 billion. (Forbes, 2013)
Thalidomide, one of the most successful repurposing drug was withdrawn due to the defects in children when the mother took the drug during the first trimester of their pregnancies. It has serendipitously found to be effective in the treatment of ENL (1964) and multiple myeloma (1999) leading to even more successful derivatives with global sales of $8.2 billion in 2017, such as lenalidomide (Revlimid, Celgene). (NRDD, 2018)
Current Practice & Challenges
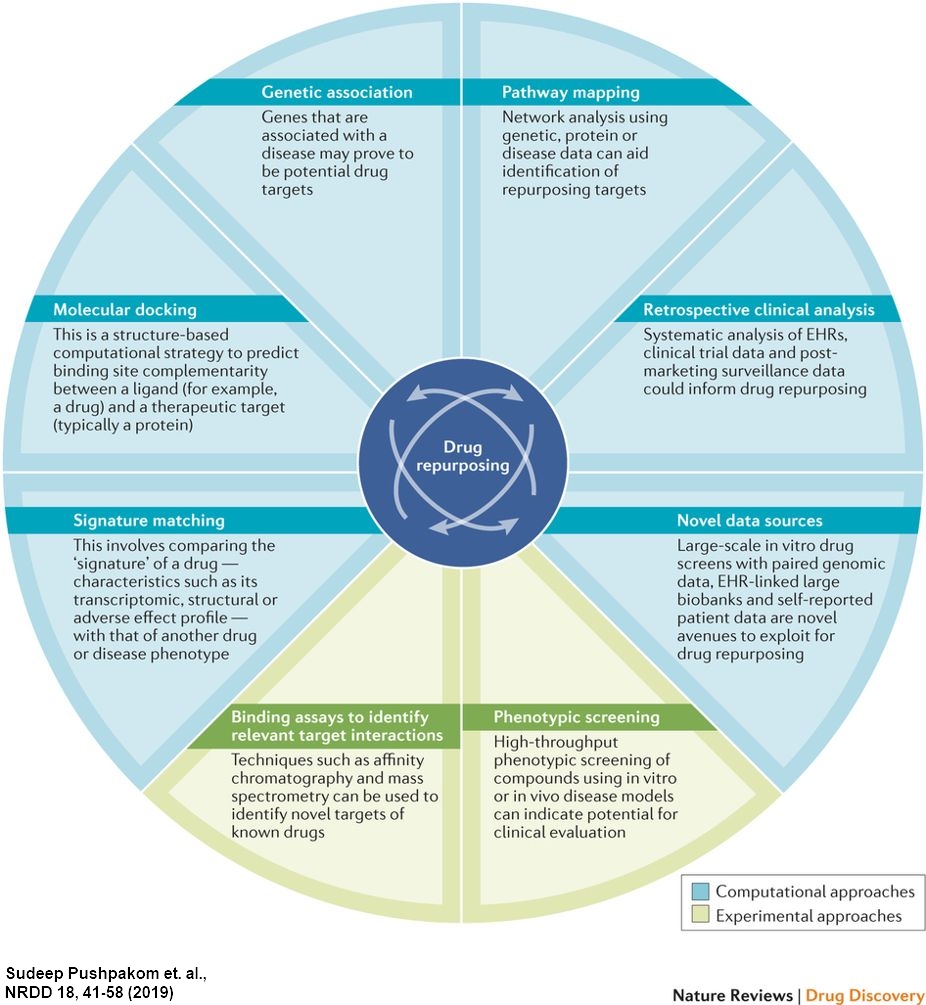
In general, a drug repurposing strategy consists of three phases: hypothesis generation, mechanistic assessment, and evaluation of efficacy. Hypothesis generation is the most critical step in this process and there are computational and experimental methods that can be used to generate hypotheses as shown on the left. (Clin Pharmacol Ther., 2013)
Computational approaches include systematic analysis of all types of data, such as gene expression, chemical/target structures. These approaches utilize feature similarities such as gene expression signatures, drug/target structural similarities, and/or molecular docking. We can also take advantage of side information similarities such as indication or adverse effects similarities. (In Silico Drug Design, 2019)
Experimental approaches include binding assays and phenotypic screening. Binding assays provide target relevance to candidate molecules, while phenotypic screening provides unbiased estimation of disease-related effects of candidate molecules. (Curr Top Med Chem, 2015)
ChallengesAdvances in biotechnology have made it possible to generate large and complex data for use in drug repurposing. However, we face a situation where we lack the ability to integrate, analyze and interpret data to improve our understanding of disease mechanisms and develop new treatment strategies. (Nature, 2015)
AI for Drug Repurposing

To meet these challeges and to improve current practice, we present Interpretable AI for Drug Repurposing for predicting drug-disease associations as well as drug-disease mechanism associations.
The main innovation of our model is the adoption and modification of current practices and recently developed deep learning models to reflect the biological structures associated with disease mechanisms in the model.
The project goals are defined as follows:
We expect that our AI model will improve current practices and become new solutions to the upcomming challenges.
SynbiDrug Repurposing Model
AI Model construction
To develop optimized AI model, we have investigated various feature combinations and DL model architectures:
- Feature Combinations
- Chemical structure: SMILES, ECFP, 2D Pharmacophores, molecular graph
- Targets: Amino-acid sequence embedding, one-hot encoding, drug-target relations
- Gene expression: total genes, landmark genes
- Cellular mechanism activities
- Side-effects
- DL models
- Deep Neural Network (DNN)
- Convolutional Neural Network (CNN)
- Long-Short Term Memory (LSTM)
- Gated Recurrent Units (GRU)
- Recurrent Convolutional Neural Network (RCNN)
- Convolution Recurrent Neural Network (CRNN)
- Graph Neural Network (GNN)
- AutoEncoder (AE)
- CapsuleNet
- Final optimized DL models
- Common Features: Drug multi-feature utilization, Error-weighted and hierarchical multimodal learning
- Drug Indication Prediction: Intercorrelating mechanism - CapsNet
- Drug Target Prediction: Classification dimension reduction - AutoEncoder
Performance
Prediction performance of drug-target interaction and drug-indication prediction was validated using public data and own experimental data (target binding assay and in vitro cellular assay).Public data validation
| Prediction | Accuracy | Sensitivity | Precision | Specificity |
|---|---|---|---|---|
| Drug-target binding | 90.3% | 81.4% | 88.1% | 99.1% |
| Drug-cancer indication | 89.2% | 87.3% | 89.8% | 93.3% |
Own experimental data validation
| Prediction | Accuracy | Sensitivity | Precision | Specificity |
|---|---|---|---|---|
| Drug-target binding | 69.8% | 75.6% | 63.7% | 57.9% |
| Drug-target activity | 65.7% | 69.8% | 42.8% | 55.7% |
| Drug-cancer indication | 69% | 73.1% | 68.1% | 79.6% |